Sunday, April 29, 2007
PCR_的經驗與感想

These days I tried so many times to get my POT1 PCR product to get reamplified by the same PCR condition used to generate itself from circular POT1 plasmid. (Fig 1. you can see lane 3 and 4 only have primer but not 1.9kb band!)
The reason why I want to do this is I cannot get the POT1 PCR digested with BamH1 ligated to the pcDNA5/HA/MCS/eGFP clone (also digested with BamH1). After awhile, I star to think about my POT1 PCR product has some problems. Therefore, I try to reamplify the POT1 PCR product.
I tried different renature time (58 to 68), different extention time (1min to 5min), dilution of my POT1 PCR product (1/10 fold).... just cannot get it work. THAT IS REALLY BOTHERING ME.....
To solve the problem, I finally sit down and think about it. Since the problem is after I got my PCR product from circular plasmid.
1. whether the PCR product has some inhibitor in it due to the PCR clean kit?
2. whether linear duplex DNA is very differnt that circular DNA like Tm to perform PCR?
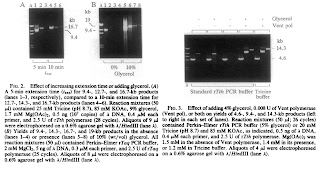
As shown in Fig.2: I digested the POT1 vector and plasmid with Pst1 (this site is in the pcDNA5/FAT plasmid not in the POT1 gene), and digested the POT1 PCR product with EcoR1 (this site is in the POT1 gene). The result shows clearly that the PCR product can be digested and the two bands add up similar to 1.9, which suggests the PCR product is really a POT1 gene and works fine. That eliminate my concern 1.
Later I performed PCR using these sample compared with original DNA samples. In addition, I tried again the previous POT1 PCR product, just want to put it as a negative control. As shown in Fig 3, surprisingly, the Pst1 digested POT1 can generate a 1.9 kb band, so the linear DNA is indeed a PCR substrate, no doubt about it! More striking is the POT1 PCR sample can also generate a 1.9 kb band this time!! (lane 5). Also, the PCR product from lane 3 & 4 can go to a second round PCR just as good as its parental ones.
So, suddently, everything turns out to be as what one would predict ! So , what is the problem for this month I 've been througn??????
One thing is differnt in this time's experiment: "The denature time increases to 3 min". After reasoning, I think this one can explain everything. The linear duplex DNA did have higher Tm compared to circular one right?.. the topology problem weight in. Although 94 degree is definitely high enough for all kind of dupelx lenght. but one thing I did not realize, it takes times to reach equilibrium. I think 2 min is not enough to denature a long duplex DNA, the Tm of primer is much lower than it (60 vs. 85). So when the temperature cool down from 94 to 62, the long duplex can replace the primer just before polymerase can access the end. Therefore, by taking longer time, the long duplex did seperate, and the time it take to aneal is more than the time Polymerase can access the primer.
啟示:
Even though , I haven't got my clone yet. But this result really stimulates me.
1. When I encounter a problem, don't try to repeat it again and again, ... THINK ABOUT IT FIRST...... This is an important attitude, a habit, a philosophy. Narrow down the question to specific points, designing practicle way to answer these questions, therefore I can test the hypothesis and increase my knowledge and experience.
2. Always keep thermodynamics and kinetics in mind, think about question by this angle. What should it be in this condition? where should it go, becomes? What does it indicate?
Monday, April 16, 2007
Competent cell by Rubicin Chloride
SO (1 liter) : tyrptone 20g
yeast extract 5g
NaCl 0.5g
KCl 2.5mM adjust pH to 7.0 (0.2ml NaOH 5M)
autoclave
SOB: add MgCl2 10mM; MgSO4 10mM
Fresh prepare:
RF1: RbCl 100mM
MnCl2.4H2O 50mM
KoAc 30mM
CaCl2.2H2O 10mM
Glycerol 15% adjust pH to 5.8 with 0.2M acetic acid
sterile by 0.2u filter
RF2: MOPS 10mM
RbCl 10mM
CaCl2.2H2O 7.5mM
Glycerol 15% adjust pH to 6.8 with NaOH
Sterile by 0.22 filter
--------------------------------------------------------------------------------
Working Medium//
125mL SO :
trypton 2.5g
yeast extract 0.025g
NaCl 0.0625g
1M KCl 312.5uL
ddH2O 125mL adjust pH to 7.0 with NaOH by testing paper//Autoclave
add 1M MaCl2 1.25mL/ 1M MgSO4 1.25mL
Fresh prepare RF1 & RF2 then in 4oC
*50mL RF1
RbCl 0.6g
MnCl2.4H2O 0.495g
KoAc 0.147g
1M CaCl2 0.5ml
50% Glycerol 15mL
ddH2O 50mL ..adjust pH to 5.8 with acetic acid by testing paper// 0.22 u filter
*10mL RF2
RbCl 0.12g
1M MOPS 0.1 ml
1M CaCl2 75uL
50% Glycerol 3ml
ddH2O 10ml .. adjust pH to 6.8 by paper// 0.22 u filter
*Competent cell
Day I: prepare fresh plate
Day II: inoculate 2-5ml SOB with 2-3 colony, 37oC overnight
Day III: inoculate o/n culture (1:100) to 125ml SOB, 270rpm. to O.D.550=0.2-0.3 (5E7cell/ml)
//following steps should be on ice//
pellet cells at 2.5k, 4oC 12-15min;
Resuspend cell in 1/3 volumn RF1, place on ice for 15 min to 2 hrs, pelet at 2.5k, 4oC 12-15min;
Resuspend cell in 1/12.5 ml volumn RF2, swirling on ice for 15min;
Aliquote 100-200ul/tube (pre-chilled on ice);
store at -70oC
Efficiency test by Fu-Kai is about 3E6 cells/ug...
Friday, April 13, 2007
Effective amplification of long targets from cloned inserts and human genomic DNA.

Reference 1:
Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5695-9.
Cheng S,Fockler C,Barnes WM,Higuchi R .
Department of Human Genetics, Roche Molecular Systems, Inc., Alameda, CA 94501
Reaction : 8% Glycerol, 2%DMSO, 0.2mM dNTP, 0.5 uM primer set, Taq 5u/pfu 0.2u mixture.
Cycle (2 steps PCR):
94oc 10 sec preheat.
94oc 10 sec (denaturation)
Tm 5-22 min (annealing and extension)
Reference 2:
Nucleic Acids Res. 1992 Sep 11;20(17):4567-73
Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR
Huang MM,Arnheim N,Goodman MF
Department of Biological Sciences, University of Southern California, Los Angeles 90089-1340
Friday, April 06, 2007
The N-end rule

Proc Natl Acad Sci U S A. 1996 Oct 29;93(22):12142-9.
The N-end rule: functions, mysteries, uses.
Varshavsky A.
Science. 1986 Oct 10;234(4773):179-86.
In vivo half-life of a protein is a function of its amino-terminal residue.
Bachmair A,Finley D,Varshavsky A.
When a chimeric gene encoding a ubiquitin-beta-galactosidase fusion protein is expressed in the yeast Saccharomyces cerevisiae, ubiquitin is cleaved off the nascent fusion protein, yielding a deubiquitinated beta-galactosidase (beta gal). With one exception (proline), this cleavage takes place regardless of the nature of the amino acid residue of beta gal at the ubiquitin-beta gal junction, thereby making it possible to expose different residues at the amino-termini of the otherwise identical beta gal proteins. The beta gal proteins thus designed have strikingly different half-lives in vivo, from more than 20 hours to less than 3 minutes, depending on the nature of the amino acid at the amino-terminus of beta gal. The set of individual amino acids can thus be ordered with respect to the half-lives that they confer on beta gal when present at its amino-terminus (the "N-end rule"). The currently known amino-terminal residues in long-lived, noncompartmentalized intracellular proteins from both prokaryotes and eukaryotes belong exclusively to the stabilizing class as predicted by the N-end rule. The function of the previously described posttranslational addition of single amino acids to protein amino-termini may also be accounted for by the N-end rule. Thus the recognition of an amino-terminal residue in a protein may mediate both the metabolic stability of the protein and the potential for regulation of its stability.
The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies.
Mogk A,Schmidt R,Bukau B (summary of the N-end rule)

Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase.

J Biol Chem. 1994 Dec 23;269(51):32678-84.
Shuman S.
Molecular Biology Program, Sloan-Kettering Institute, New York, New York 10021.
Construction of chimaeric DNA molecules in vitro relies traditionally on two enzymatic steps catalyzed by separate protein components. Site-specific restriction endonucleases are used to generate linear DNAs with defined termini that can then be joined covalently at their ends via the action of DNA ligase. A novel approach to the synthesis of recombinant DNAs exploits the ability of a single enzyme, vaccinia DNA topoisomerase, to both cleave and rejoin DNA strands with extreme specificity at each step. Placement of the CCCTT cleavage motif for vaccinia topoisomerase near the end of a duplex DNA permits efficient generation of a stable, highly recombinogenic protein-DNA adduct that can religate only to acceptor DNAs that contain complementary single-strand extensions. Linear DNAs containing CCCTT cleavage sites at both ends (bivalent substrates) can be activated by topoisomerase and inserted into a plasmid vector in a simple and rapid in vitro procedure that is especially well suited to the molecular cloning of polymerase chain reaction-amplified DNAs. Activation of polyvalent (e.g. branched) DNA substrates by topoisomerase offers a potentially powerful method for the synthesis of two- and three-dimensional polynucleotide networks.
Thursday, April 05, 2007
A Luminol/Iodophenol Chemiluminescent Detection System for Western Immunoblots

 Analytical Biochemistry
Analytical Biochemistry Volume 258, Issue 1 , 10 April 1998, Pages 146-149
Alexander F. Yakunin1 and Patrick C. Hallenbeck
Département de microbiologie et immunologie, Université de Montréal, Montréal, Québec, H3C 3J7, Canada
Solution 1: 1mM Luminol, 8 mM 4iodophenol, 50mM glycin-NaOH (pH 9.6)
Solution 2: 17.6 mM H2O2
mix Sol.1 & Sol.2 before reaction.

